Welcome to Single Cell Expression Atlas help
Data Summary and the developmental tree construct
At the time of this publication, the database contains 10 datasets covering 18,413 single-cells and 176 cell groups. According to the notation of the data resources, we classified these cell groups into 35 developmental stages. Every mammalian individual is developed from the totipotent zygote. Mammalian pre-implantation development is a complex process including a series of cell divisions from 1 cell to 2 cells, 2 cells to 4 cells, 4 cells to 8 cells, 8 cells to 16 cells and 16 cells to blastocyst. After that, nearly all of the human tissues are original from embryoblast. Then a developmental tree was constructed based on the development process of the multicellular organism(Figure 1). The detailed cell number in each stage is shown in Figure 2 and the datasets summary is available at "Data Summary" page.
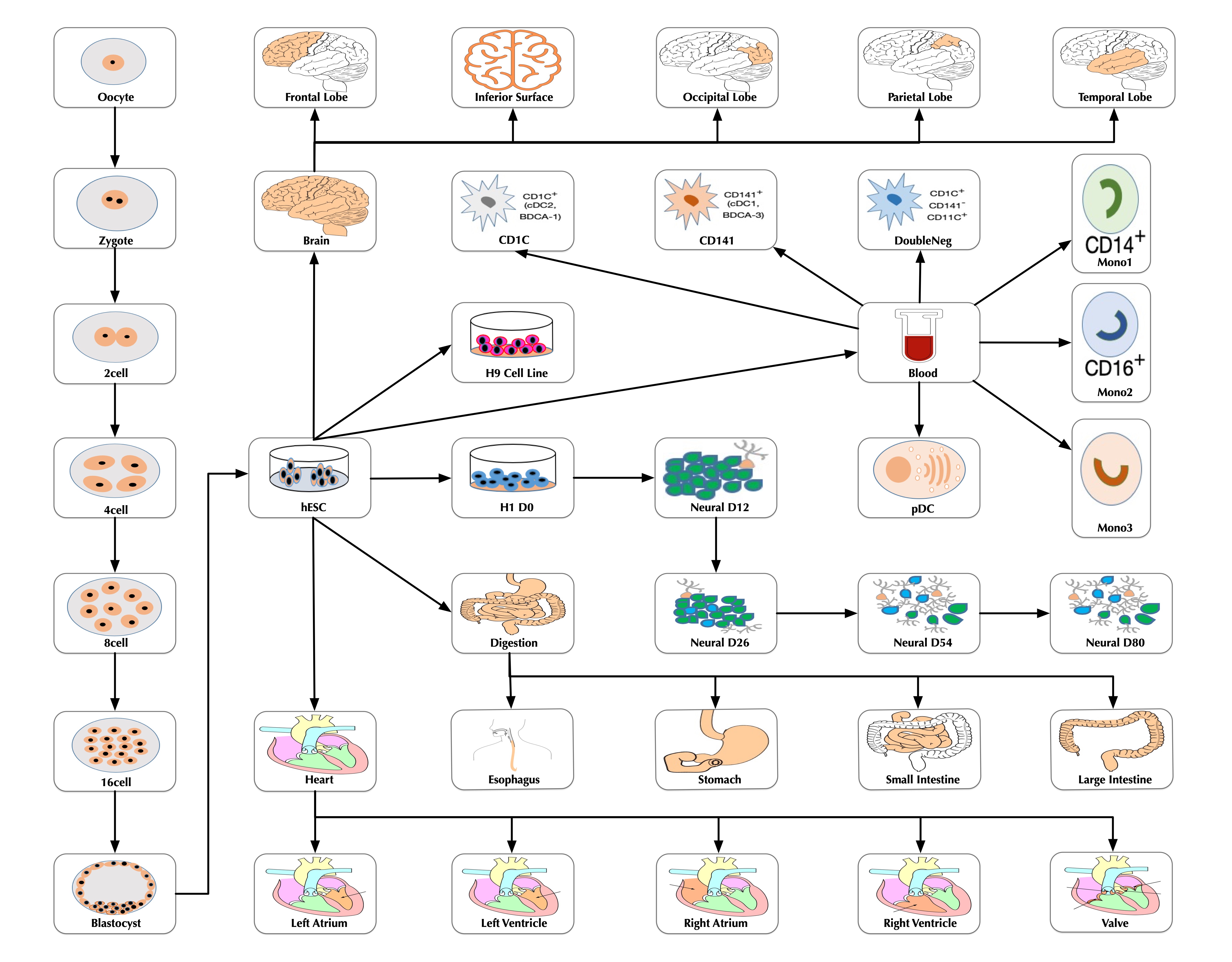
Figure 1: The developmental tree.
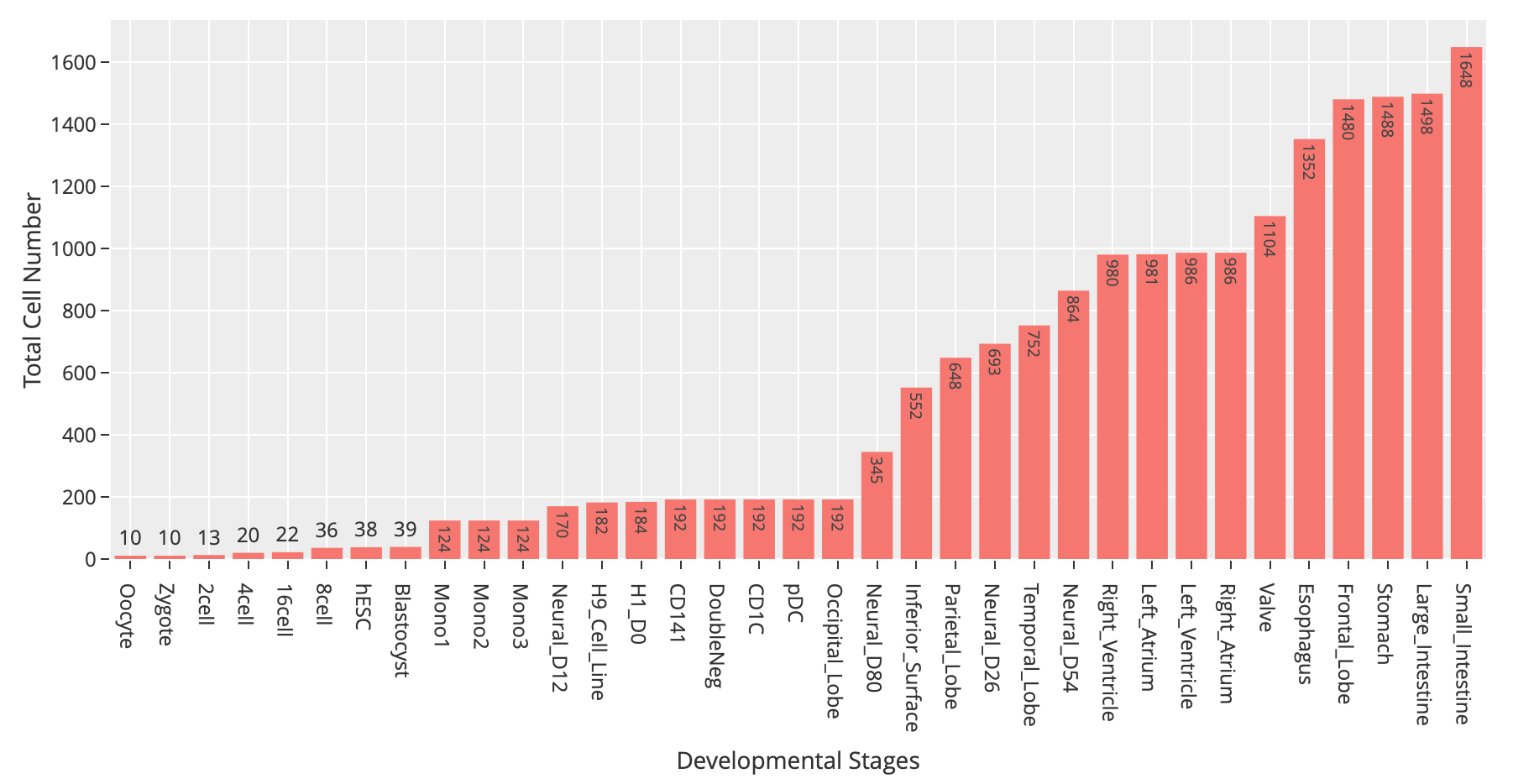
Figure 2: Statistics of cell numbers of 35 developmental stages.
How to search the gene expression
Here you can use the Gene search box to search your interested gene expression in different development stages. You may search its gene ID via its gene symbol (e.g., MYL2) or Ensembl ID (e.g., ENSG00000127837)(Figure 1A).
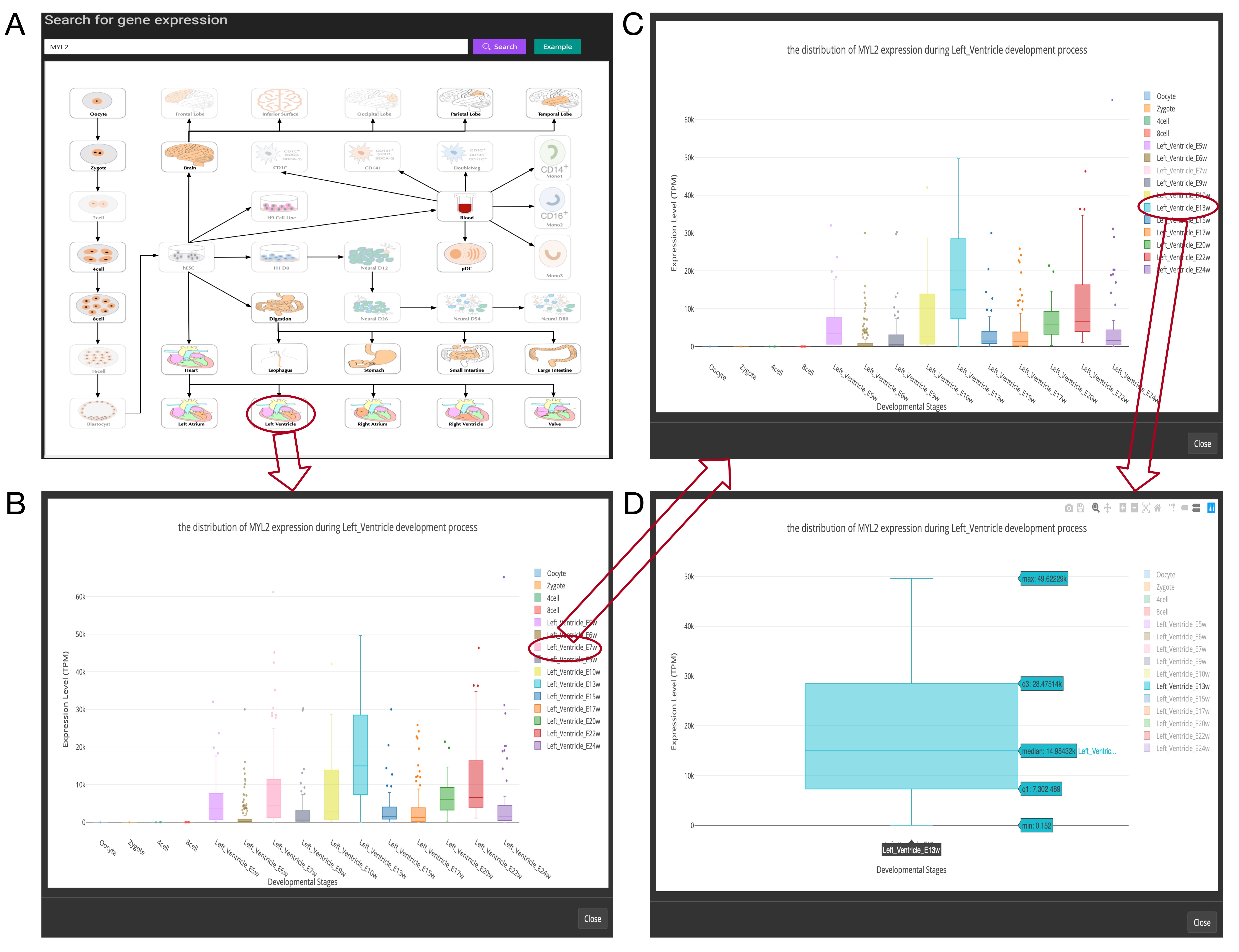
Figure 3: Overview of the gene expression search page. (A). Searching result of the gene “MYL2”. (B). Boxplot shows expression level distribution of MYL2 during developmental process by clicking the image. (C). The function of removing uninterested developmental stages by clicking the name of the stage listed in the figure legend. (D). An example of double-clicking on a stage name.
Gene expression search result
The searching result will be displayed in the developmental tree. Specifically, if the searched gene (“MYL2” gene as an example) is not expressed at one stage, the stage image will be disabled and cannot be clicked (the light-colored images in Figure 3A). Furthermore, the interactive boxplot of gene expression level along with a selected developmental pathway is available by clicking the stage image (Figure 3B). To illustrate the interactive function of this boxplot, we took the distribution of the MYL2 expression during right ventricle process as an example. Clicking on the stage name “Left_Ventricle_E7w” listed in the graph legend can remove the boxplot data of this stage (Figure 3C). This function allows users to compare their interested stages. Moreover, double- clicking on a stage name allows users to view detail gene expression value of this stage (Figure 3D). These boxplots can be download in PNG format for further usage.
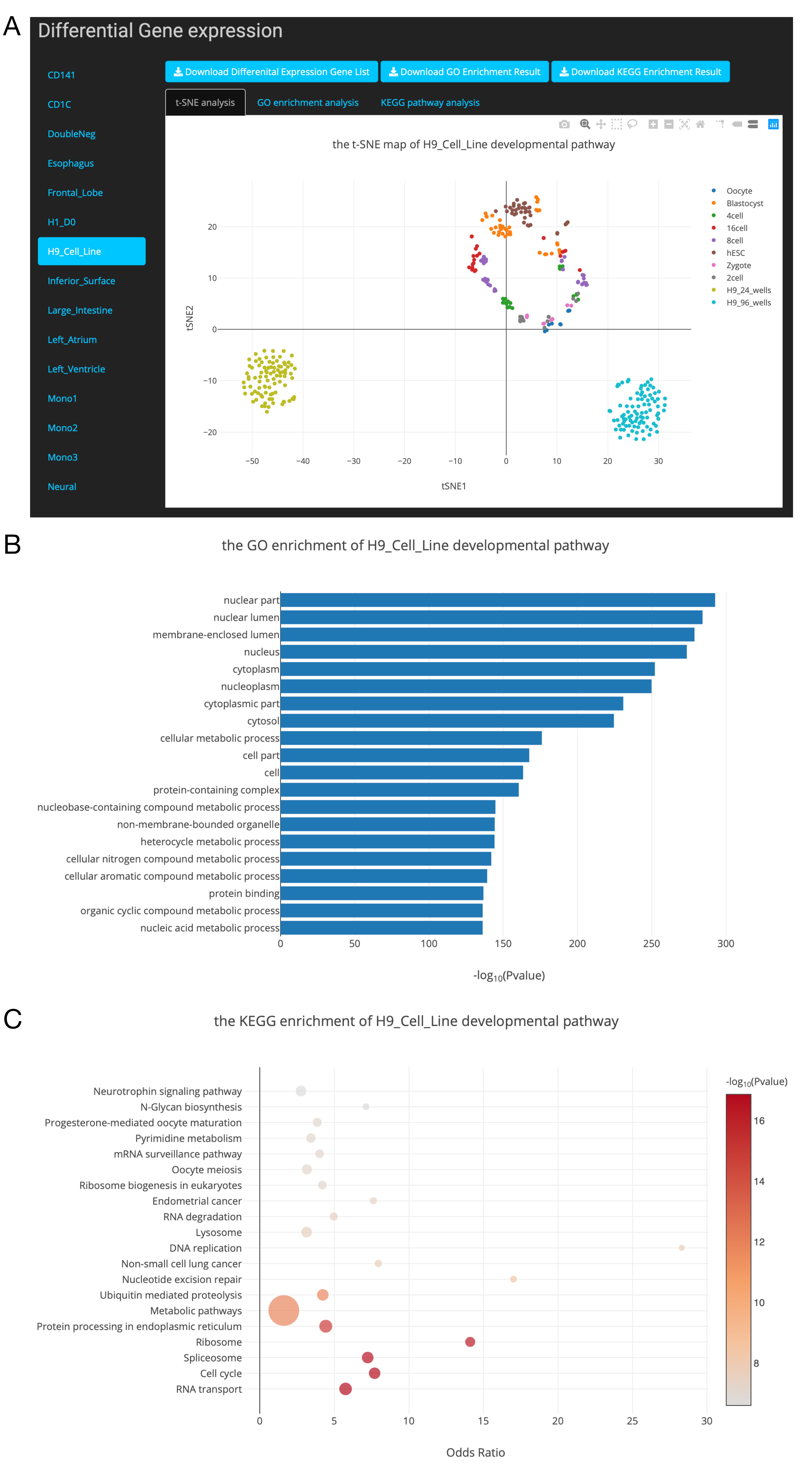
Differential gene list collection for 24 developmental pathways
In this study, we performed differentially expressed (DE) gene analysis for 24 developmental pathways. Finally, 24 differential gene lists were collected into the SCDevDB. Users can download these gene lists by clicking the Download button in Differential Gene List Collection page. Moreover, we performed t-SNE analysis using these differential gene lists and the result is displayed using an interactive scatterplot (Figure 4A). Subsequently, GO and KEGG enrichment analysis of the DE genes were performed using R packages and top 20 significantly enriched GO or KEGG terms were selected to show potential functions of these DE genes (Figure 4B, 4C). In addition, tables showing all of the GO or KEGG terms are also available and free to download on the “Differential Gene List Collection” page. These scatterplots and bar chart can be download in PNG format for further usage.